3D mapping of loops in human genome reveals structure and hidden switches
Researchers have assembled the first high-resolution, 3D maps of entire folded genomes which show how a "genomic origami" or folding allows the same genome to produce different types of cells.
The five-year project to identify the loops in the human genome was a collaboration between researchers at Harvard University, Baylor College of Medicine, Rice University and the Broad Institute of Harvard and MIT.
The team collected billions of snippets of DNA that were analysed for signs of loops. They found that loops and other genome folding patterns are an essential part of genetic regulation.
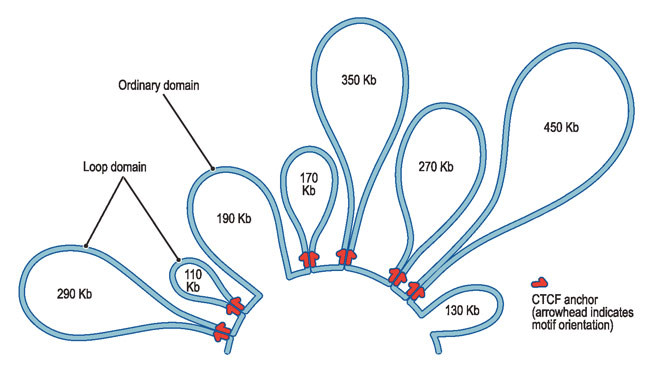
Loops form when two bits of DNA that are far apart in the genome sequence end up in close contact in the folded version of the genome in a cell's nucleus.
The maps of looping reveal thousands of hidden switches, knowing which helps scientists studying genes that can cause cancer or other diseases.
"More and more, we're realising that folding is regulation," said study co-first author Suhas Rao, a researcher at Baylor's Center for Genome Architecture and a 2012 graduate of Harvard College. "When you see genes turn on or off, what lies behind that is a change in folding. It's a different way of thinking about how cells work."
The largest loops in the genome are in women, with some gigantic loops almost up to 30 times the size of what is seen in males.
They found that many of the loops present in humans are also present in mice, implying that these specific folds have been preserved over nearly one hundred million years of evolution.
The Hi-C methodology for sequencing genomes in 3D introduced five years ago had to be overhauled and the team also faced significant computational hurdles.
"In 2009, we were dividing the genome into 1-million-base blocks, and here we are dividing it into 1,000-base blocks," said Miriam Huntley, a doctoral student at Harvard.
"Since any block can collide with any other block, we end up with a problem that is a millionfold more complicated. The overall database is simply vast."
New methods were also developed to speed up the data processing and reduce experimental noise and irregular fluctuations that tend to obscure weak signals in the data.
In the process, the team discovered a series of rules about how and where loops can form in the genome.
"If DNA were a shoestring, you could make a loop anywhere. But within the cell, the formation of loops is highly constrained," said Rao.
"The loops we see almost all span fewer than 2 million genetic letters; they rarely overlap; and they are almost always associated with a single protein, called CTCF."
A stunning discovery was about how CTCF proteins form a loop pointing at each other, however far they were, forming a genomic yin and yang, as the team notes.
The research appears online in Cell.
© Copyright IBTimes 2025. All rights reserved.





















